Deep Learning is able to change many things in our world. We can apply it in many ways. Vera presents us medical applications to image classification. Will Machine Learning exceed humans on lesion detection ?
Introduction
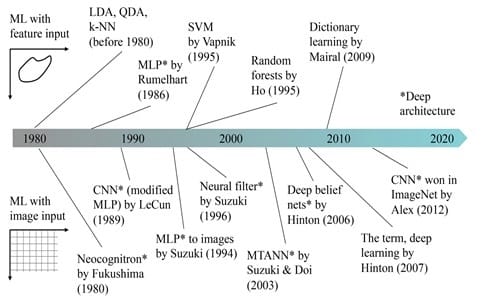
In the last few years, deep learning (DL) has become very popular in so many domains thanks to an event that took place in 2012. A DL approach named AlexNet [1], based on Convolutional Neural Networks (CNN) designed by Alex Krizhevsky, won the ImageNet Large Scale Visual Recognition Challenge (ILSVRC) [2], the world’s best-known computer vision competition. The error rate was 15.3 %, almost 11 % lower than its second runner up’s (26 %) [3]. As a result, this achievement was considered by the MIT Technology Review as one of the top 10 breakthrough technologies of 2013. This event marked a milestone for the DL world, where researchers all over the globe have started showing great interest in applying DL in a variety of fields.
It is important to note that Hinton proposed the term Deep Learning in 2007 for Machine Learning models with a high-level object representation, but the naming was not recognized until 2012.
In the medical field, Machine Learning (ML) is mainly used in Computer-Aided Diagnosis (CAD) and medical image analysis for classification (lesion or non-lesion, benign or malignant). Features such as textural measures, shape descriptors and contour representations are used by ML in order to optimally separate the different classes. This type of ML is known as feature-based ML.
DL, on the other hand, does not require object segmentation and feature calculation. DL is actually known as end-to-end ML because it takes in images as inputs and yields classification results as outputs. Normally, DL involves four or more layers in order to meet its classification objectives [4]. In fact, DL networks would as well be called « ML with image input or image-based ML ». This same network can be called « deep image-based ML » or « deep feature-based ML » when the architecture is deep. The difference between feature-based ML and image-based ML is shown in Figure 1. It is important to note that ML and DL can also be used to study data other than images, however the scope of this article only covers their application on medical images.
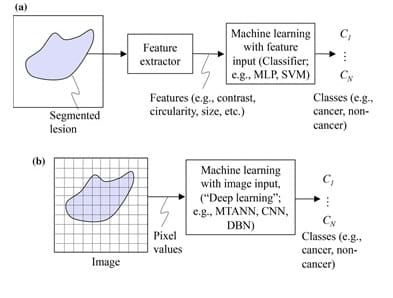
Despite the fact that the subject of segmentation has occupied researchers for a very long time, it remains a difficult task to segment objects with complex structures or with complicated backgrounds. Besides, feature definition and extraction is a real challenge, for those features might not always be reliably discriminative. Therefore, we can see that deep learning, with its ability to perform classification without the need for segmentation and feature extraction, can provide a higher performance for the classification of complex images. However, even if DL is widely appreciated for its powerful performance, a final validation by human experts is still a necessary step. This is in addition to the vast amounts of open-source data available online that might result in a lower DL performance due to its poor quality at times.
A historical overview of ML methods in medical imaging is presented in Figure 2. Before DL became very popular starting 2013, feature-based ML was being widely used. Ever since the DL boom in 2013, image-based ML became extremely popular.
This article will discuss DL in medical imaging: The transformation from ML to DL and the birth of the massive-training artificial neural network (MTANN) [5].
MTANN
Another type of ML that differs from the feature-based ML was introduced in CAD applications under the name of Massive-Training Artificial Neural Network (MTANN) by Suzuki et al. [6]. Images serve as inputs to MTANN instead of features. Therefore, this type of ML is known as image-based ML. Among other applications, MTANNs were used to reduce false positives in lung-nodule detection, to classify them as benign or malignant, to separate soft tissue from bone, etc. As a result, MTANN is an end-to-end ML process that takes in an image and yields the classification result together with the detected object without the need for segmentation and feature extraction.
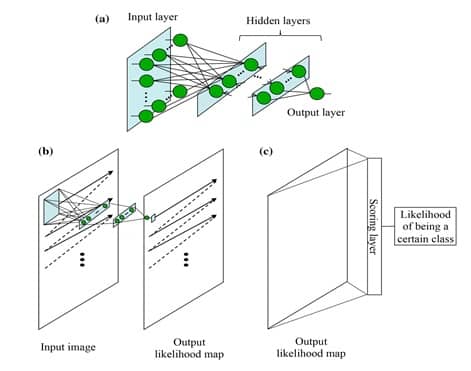
The MTANN architecture is shown in Figure 3-a. It consists of an input layer, multiple hidden layers, and an output layer. The input to the MTANN is an image patch or a subregion of pixels taken from the original image of interest. The activation functions employed in the hidden layers are sigmoid functions in case of non-linear processing. A linear function is used instead of a sigmoid function to serve as an activation function in the output layer. MTANN convolves or scans using convolution the input image patch on the full-size original input image, as shown in image 3-b. A continuous scalar value is yielded as an output, associated to the center pixel of the fed image patch. Therefore, the final MTANN output is a likelihood map with local pixel-level information, which is not the case of CNN.
In order to give to MTANN the classification functionality of CNN, an additional layer named « scoring layer » is to be added after the current output layer. The scoring layer converts the likelihood map into a single unique score of belonging likelihood. This scoring layer is illustrated in Figure 3-c.
After MTANN is trained, it will output the maximum value when an object of interest is situated in the center of the fed subregion. The values become lower farther from the center, and they eventually turn to 0 outside the object.
MTANN vs CNN
CNN takes an image as an input and yields the image’s class as an output, such as benign or malignant. The CNN architecture is shown in Figure 4.
MTANNs and CNNs are both in the category of image-based ML or DL. However, there are three major differences between the two networks :
In CNN, convolution takes place inside the network (Figure 5) while it happens outside the network in MTANN (Figure 4).
CNN’s output is normally a class, whereas MTANN outputs an image (a map of continuous values).
Due to an elevated number of necessary model parameters, CNN requires a great number of images for training (can be as high as 1 million). MTANN, on the other hand, needs only a few images for training (±10). With a GPU implementation, CNN usually needs several days, whereas MTANN only takes a few hours.
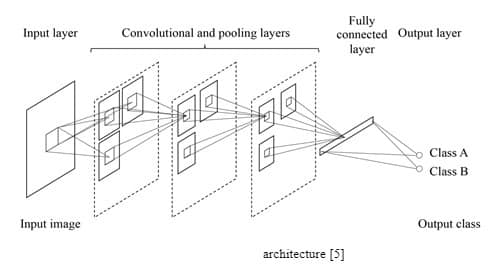
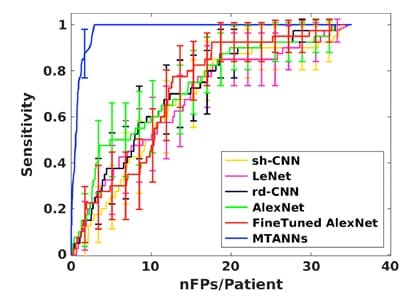
Figure 5 shows a comparison between MTANN and well-know CNNs (shallow CNN (sh-CNN), LeNet, relatively deep CNN (rd-CNN), AlexNet, and FineTuned AlexNet) [7]. This comparative study was conducted to detect and classify lung nodules. It clearly shows the outstanding performance of MTANN for experiments carried out under the same conditions. However, it is important to note that when the training image database was increased (only for CNNs), these networks performed better, but the difference remained significant.
MTANN Applications
MTANN has been used in several medical applications to increase diagnostic precision and to provide a better prognosis. Some of these applications are discussed in what follows.
1. Lesion classification:
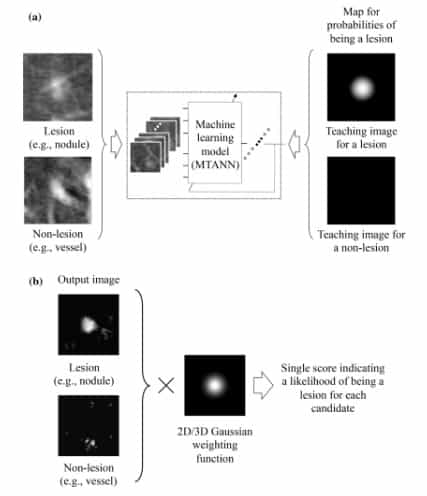
One application of MTANN in medical imaging is the differentiation between lesions and non-lesions, as shown in Figure 6. In order to be able to enhance lesions and suppress non-lesions, MTANN is trained using an image with a map of value distributions for probable lesions [5].
Another example is shown in Figure 7. A lung nodule is shown with an arrow in Figure 7 (a), and MTANN’s success in enhancing this sick region only and suppressing the healthy remaining of the chest is clearly displayed in Figure 7 (b) [8].
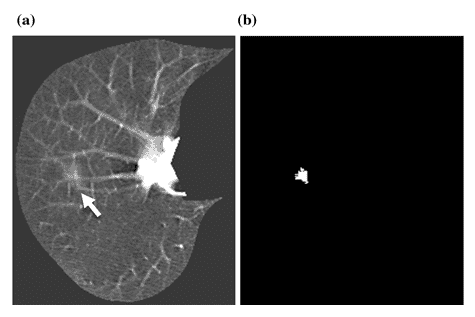
Figure 7 : (a) A lung nodule is indicated with the white arrow. (b) MTANN’s result for nodule enhancement [8]
2. Segmentation
Even though segmentation is quite challenging sometimes due to unclear and fuzzy borders of the organs (Figure 8 (a)), the predecessors of MTANN, named Neural Edge Enhancers (NEE), were able to enhance the edges of organs, in this case the left ventricle (Figure 8 (b)). The closeness of the segmentation results by both a cardiologist and NEE can be easily seen in Figure 8 (c) [9]
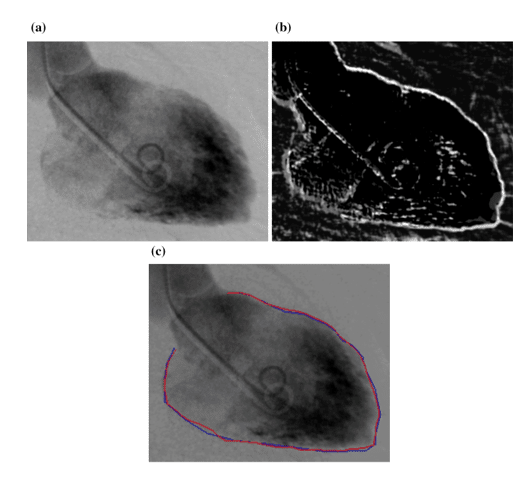
Figure 8 : (a) The image of a left ventricle of the heart (b) Edge enhancement by NEE (c) Comparison between hand segmentation and NEE results [9]
3. Bone – soft tissue separation
According to studies, 82 – 95 % of undetected lung cancers are due to overlying bones [10], such as the ribs. MTANN was developed to separate bones from soft tissues, as shown in Figure 9 [11].
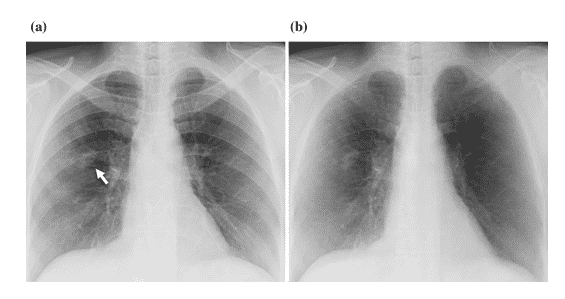
Figure 9: (a) Lung X-Ray with a nodule shown by the white arrow (b) The results of MTANN application to separate bone form soft tissue [11]
Conclusion
As a summary, feature-based ML was dominating before DL was introduced. DL made it possible to take images directly as inputs without undergoing segmentation or feature extraction. This fact about DL together with its deep network reduced uncertainty of classification and object detection.
DL is an extremely promising and a very fast-growing field. Medical imaging is being led to yet another whole new level of advancement and high performance thanks to DL. It is predicted that medical imaging will use DL as its mainstream technology in the near future and for many decades to come.
References
[1] http://papers.nips.cc/paper/4824-imagenet-classification-with-deep-convolutional-neural-networks
[2] http://www.image-net.org/challenges/LSVRC/2012/
[3] http://www.image-net.org/challenges/LSVRC/2012/results.html
[4] https://meritis.fr/ia/deep-learning/
[5] Suzuki, Kenji. « Overview of deep learning in medical imaging. » Radiological physics and technology 10.3 (2017): 257-273.
[6] Suzuki, Kenji, et al. « Massive training artificial neural network (MTANN) for reduction of false positives in computerized detection of lung nodules in low‐dose computed tomography. » Medical physics 30.7 (2003): 1602-1617.
[7] Tajbakhsh, Nima, and Kenji Suzuki. « Comparing two classes of end-to-end machine-learning models in lung nodule detection and classification: MTANNs vs. CNNs. » Pattern recognition 63 (2017): 476-486
[8] Suzuki, Kenji. « A supervised ‘lesion-enhancement’ filter by use of a massive-training artificial neural network (MTANN) in computer-aided diagnosis (CAD). » Physics in Medicine & Biology54.18 (2009): S31.
[9] Suzuki, Kenji, et al. « Extraction of left ventricular contours from left ventriculograms by means of a neural edge detector. » IEEE Transactions on Medical Imaging 23.3 (2004): 330-339. ,
[10] Austin, J. H., B. M. Romney, and L. S. Goldsmith. « Missed bronchogenic carcinoma: radiographic findings in 27 patients with a potentially resectable lesion evident in retrospect. » Radiology 182.1 (1992): 115-122.
[11] Suzuki, Kenji, et al. « Suppression of the contrast of ribs in chest radiographs by means of massive training artificial neural network. » Medical Imaging 2004: Image Processing. Vol. 5370. International Society for Optics and Photonics, 2004.



Pas encore de commentaires